Enlarge (credit: University of Washington )
Thanks to the development of DNA-sequencing technology, it has become trivial to obtain the sequence of bases that encode a protein and translate that to the sequence of amino acids that make up the protein. But from there, we often end up stuck. The actual function of the protein is only indirectly by its sequence. Instead, the sequence dictates how the amino acid chain folds and flexes in three-dimensional space, forming a specific structure. That structure is typically what dictates the function from the protein, but obtaining it can require years of lab work.
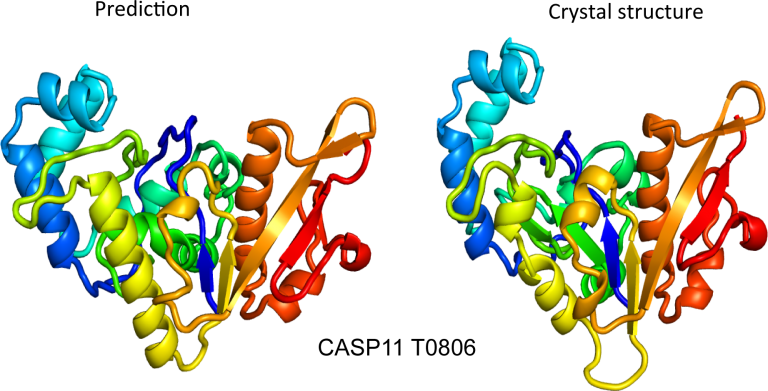
For decades, researchers have tried to develop software that will can take a sequence of amino acids and accurately predict the structure it will form. Despite this being a matter associated with chemistry and thermodynamics, we’ve only had limited success—until last year. That’s when Google’s DeepMind AI group announced the existence of AlphaFold , which can typically predict structures with a high degree of accuracy.
At the time, DeepMind said it would give everyone the details on its breakthrough within a future peer-reviewed paper, which usually it finally released yesterday. In the meantime, some academic researchers got tired of waiting, took some of DeepMind’s insights, and made their own. The paper describing that effort also was released yesterday.