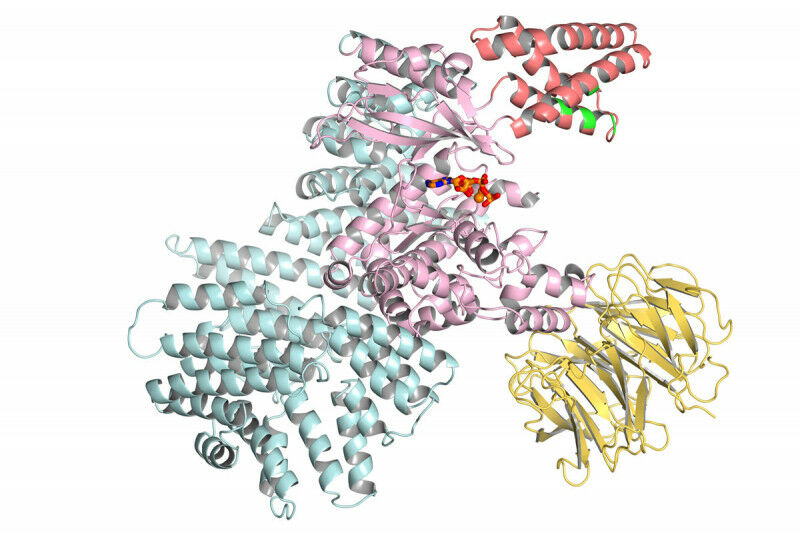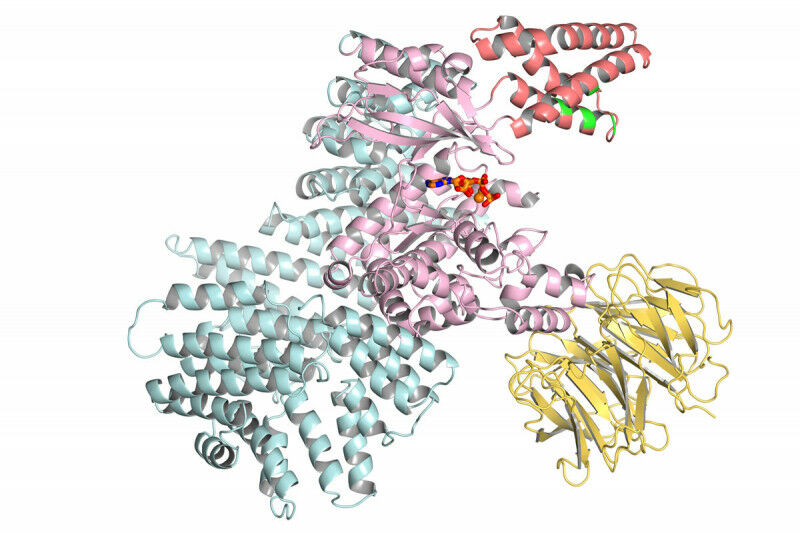
Enlarge (credit: Sloan-Kettering)
Just one week after Google’s DeepMind AI group finally described its biology efforts in detail, the company is releasing a paper that explains how it analyzed nearly every protein encoded in the human genome and predicted its likely three-dimensional structure—a structure that can be critical for understanding disease and designing treatments. In the very near future, all of these structures will be released under a Creative Commons license via the European Bioinformatics Institute, which already hosts a major database of protein structures.
In a press conference associated with the paper’s release, DeepMind’s Demis Hassabis made clear that the company isn’t stopping there. In addition to the work described in the paper, the company will release structural predictions for the genomes of 20 major research organisms, from yeast to fruit flies to mice. In total, the database launch will include roughly 350,000 protein structures.
What’s in a structure?
We just described DeepMind’s software last week, so we won’t go into much detail here. The effort is an AI-based system trained on the structure of existing proteins that had been determined (often laboriously) through laboratory experiments. The system uses that training, plus information it obtains from families of proteins related by evolution, to predict how a protein’s chain of amino acids folds up in three-dimensional space.